Setting up an electronic minimization: Difference between revisions
| (21 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
Setting up an [[:Category:Electronic minimization |electronic minimization]] calculation using density-functional theory requires a few steps. The [[:Category:Input files | input files]] must be created or copied into the execution folder. This includes making a few choices for the [[KPOINTS | '''k'''-point]] sampling and [[ISMEAR | electronic smearing]], [[ALGO | minimization algorithm]], and [[:Category:Exchange-correlation functionals|exchange-correlation functionals]]. A [[Command-line_arguments#--dry-run_/_-n | dry-run]] can be used to review settings and select appropriate [[:Category:Parallelization | parallelization]] tags. After running the calculation, the | Setting up an [[:Category:Electronic minimization |electronic minimization]] calculation using density-functional theory requires a few steps. The [[:Category:Input files | input files]] must be created or copied into the execution folder. This includes making a few choices for the [[KPOINTS | '''k'''-point]] sampling and [[ISMEAR | electronic smearing]], [[ALGO | minimization algorithm]], and [[:Category:Exchange-correlation functionals|exchange-correlation functionals]]. A [[Command-line_arguments#--dry-run_/_-n | dry-run]] can be used to review settings and select appropriate [[:Category:Parallelization | parallelization]] tags. After running the calculation, the output can be analyzed. | ||
== Step-by-step instructions == | == Step-by-step instructions == | ||
=== Create the input files === | |||
'''Step | '''Step 1''': Create a {{FILE| POSCAR}} file containing the structure for which you want to compute the electronic groundstate. External tools like VESTA{{cite|vesta}}, or Python packages like the Atomic Simulation Environment (ASE){{cite|ase}} or pymatgen{{cite|pymatgen}} can help with this step. | ||
'''Step | '''Step 2''': [[:Category:Exchange-correlation_functionals#Which_exchange-correlation_method_to_choose?|Choose an exchange-correlation (XC) functional]] appropriate for your material and quantity of interest. | ||
'''Step 4''': Create a {{FILE| KPOINTS}} file to define the integration mesh in reciprocal space. | '''Step 3''': Create a suitable {{FILE|POTCAR}} file by following the instructions on our [[Preparing a POTCAR | preparing a POTCAR]] page. | ||
'''Step 4''': Create a {{FILE| KPOINTS}} file to define the [[Integrating over all orbitals|integration mesh in reciprocal space]]. Including a single k point at the origin, i.e. the Gamma point, neglects all interactions beyond the unit cell. This is appropriate for isolates systems like a single molecule or in large supercells. For bulk systems, start with a [[KPOINTS#Regular_k-point_mesh|regular mesh]]. For shorter lattice vectors, more k points are required to achieve the same sampling density. Consult the [[KPOINTS#Symmetry_reduction_of_the_mesh | symmetry reduction section]] of the {{FILE| KPOINTS}} page to select the appropriate mesh type. Alternatively to a {{FILE| KPOINTS}} file, the {{TAG| KSPACING}} can be used. | |||
'''Step 5''': Write an {{FILE| INCAR}} file. It is recommended to start from a rather minimal file, and only specify the most important tags: | '''Step 5''': Write an {{FILE| INCAR}} file. It is recommended to start from a rather minimal file, and only specify the most important tags: | ||
:* [[XC]] to specify the [[:Category:Exchange-correlation_functionals| exchange-correlation functional]] | :* [[XC]] to specify the [[:Category:Exchange-correlation_functionals| exchange-correlation functional]]. | ||
:* [[ALGO]] to select the algorithm for [[:Category:Electronic minimization |electronic minimization]]. | :* [[ALGO]] to select the algorithm for [[:Category:Electronic minimization |electronic minimization]]. | ||
:* [[ISMEAR]] to select the type of [[Smearing technique|electronic smearing technique]]. | :* [[ISMEAR]] to select the type of [[Smearing technique|electronic smearing technique]]. | ||
:* [[SIGMA]] to choose an appropriate smearing width of the [[Smearing technique|electronic smearing]]. | :* [[SIGMA]] to choose an appropriate smearing width of the [[Smearing technique|electronic smearing]]. | ||
:* [[ENCUT]] to set the plane-wave energy cutoff | :* [[ENCUT]] to set the plane-wave energy cutoff. | ||
:* [[EDIFF]] to specify the global break condition for the electronic self-consistent loop | :* [[EDIFF]] to specify the global break condition for the electronic self-consistent loop | ||
'''Step 6''' (optional): Select the appropriate version of the VASP executable. I.e. <code>vasp_gam</code> if you only want to use the Gamma point for reciprocal space integration, <code>vasp_ncl</code> for [[:Category:Noncollinear_magnetism| noncollinear]] calculations, or <code>vasp_std</code> for anything else. Then Run a [[Command-line_arguments#--dry-run_/_-n | dry-run]] calculation | === Optimize your settings === | ||
'''Step 6''' (optional): Select the appropriate version of the VASP executable. I.e. <code>vasp_gam</code> if you only want to use the Gamma point for reciprocal space integration, <code>vasp_ncl</code> for [[:Category:Noncollinear_magnetism| noncollinear magnetic]] calculations, or <code>vasp_std</code> for anything else. Then Run a [[Command-line_arguments#--dry-run_/_-n | dry-run]] calculation to validate settings and uncover possible errors. Open a terminal, go to the calculation directory that contains all input files and run | |||
/path/to/your/vasp_std --dry-run | |||
'''Step 7''' (optional): Inspect the {{FILE| OUTCAR}} file of your [[Command-line_arguments#--dry-run_/_-n | dry-run]]. Take note of the number of bands, {{TAG|NBANDS}}, and the number of '''k'''-points, NKPTS, especially. Follow the guidelines on the [[Optimizing_the_parallelization | optimizing the parallelization]] page to set {{TAG|NCORE}} and/or {{TAG|KPAR}} in the {{FILE|INCAR}} file. | |||
=== Run the calculation === | |||
'''Step 8''': Run the calculation. If you are new to VASP, or unsure about the calculation setup, run a small calculation and monitor the [[stdout | screen output]]. For parallel execution on 4 MPI ranks, the command reads | |||
mpirun -np 4 /path/to/your/vasp_std | |||
At an HPC center, submit your job with a submission script. Ask your system administrator for help. | |||
Once the calculation is finished, you have access to the [[:Category:Electronic ground-state properties| electronic ground-state properties]] via the [[:Category:Output_files| output files]] for the selected parameters. Check the {{FILE| OUTCAR}} file for warnings or advice. For help, consult the page about [[Troubleshooting_electronic_convergence| troubleshooting electronic convergence]] and search our [https://www.vasp.at/forum/| Forum] for similar issues. | |||
<div id="Step 9" style="display:inline;">'''Step 9''' (convergence study): Repeat Steps 4 - 8 with increasingly accurate parameter settings, e.g. higher cutoff energy and denser k-points mesh, and monitor your quantity of interest. Stop if the quantity of interest reaches the desired accuracy.</div> | |||
== Recommendations and advice == | == Recommendations and advice == | ||
{{NB|mind|Make sure to specify the lattice vectors and ionic positions in the {{FILE|POSCAR}} with at least 7 digits of precision to ensure the [[POSCAR#Precision_and_symmetry|symmetry analysis]] can | {{NB|mind|Make sure to specify the lattice vectors and ionic positions in the {{FILE|POSCAR}} with at least 7 digits of precision to ensure the [[POSCAR#Precision_and_symmetry|symmetry analysis]] can function accurately.}} | ||
{{NB|tip|Add only necessary tags to your {{FILE|INCAR}} file. Cluttered input is a common source of mismatched settings. | {{NB|tip| | ||
*Add only necessary tags to your {{FILE|INCAR}} file. Cluttered input is a common source of mismatched settings. | |||
{{ | *A larger smearing width {{TAG|SIGMA}} might be required to converge the calculation if your {{FILE| KPOINTS}} mesh is sparse. | ||
*{{TAG|ENCUT}} defaults to the largest [[ENMAX]] value found in the {{FILE| POTCAR}} file. Still, it is always a good idea to include it in the {{FILE| INCAR}} file to ensure comparability between different calculations. | |||
*Use the [[Command-line_arguments#--dry-run_/_-n| dry-run]] command-line argument or {{TAGO|ALGO|None}} to check the feasibility of your settings and [[Optimizing_the_parallelization| optimize parallelization tags]], without wasting computational resources. | |||
*Some warnings are a bit hidden in the [[Screen_output#The_header| header]] section of the [[Screen_output| screen output]]. Redirecting the screen output to a file and saving it can simplify troubleshooting significantly.||Tips}} | |||
== Example == | == Example == | ||
We will do a small DFT calculation of GaAs in the zincblende structure, using the local-density approximation (LDA) with the Perdew-Zunger parametrization of Ceperley-Alder Monte Carlo correlation data.{{cite|ceperley1980}}{{cite|perdewzunger1981}}. Thus our [[:Category:Exchange-correlation_functionals| XC functional]] will be set to {{TAGO|XC|CA}}. | We will do a small DFT calculation of GaAs in the zincblende structure, using the local-density approximation (LDA) with the Perdew-Zunger parametrization of Ceperley-Alder Monte Carlo correlation data.{{cite|ceperley1980}}{{cite|perdewzunger1981}}. Thus, our [[:Category:Exchange-correlation_functionals| XC functional]] will be set to {{TAGO|XC|CA}}. | ||
=== Setting up the POSCAR file === | === Setting up the {{FILE|POSCAR}} file === | ||
The {{FILE|POSCAR}} file starts with a comment line | The {{FILE|POSCAR}} file starts with a comment line and a scaling factor, which in our case corresponds to the lattice parameter of GaAs, around 5.65 Angstrom. | ||
Zincblende GaAs | Zincblende GaAs | ||
5.65000000000 | 5.65000000000 | ||
| Line 46: | Line 64: | ||
0.5000000000000000 0.0000000000000000 0.5000000000000000 | 0.5000000000000000 0.0000000000000000 0.5000000000000000 | ||
0.5000000000000000 0.5000000000000000 0.0000000000000000 | 0.5000000000000000 0.5000000000000000 0.0000000000000000 | ||
Now we define the ion types, and in the line below the | Now, we define the ion types, and in the line below the number of ions in the structure for each type: | ||
Ga As | Ga As | ||
1 1 | 1 1 | ||
Specify the positions of the atoms in direct coordinates, with Ga at the origin and As a quarter along the diagonal of the cube: | |||
Direct | Direct | ||
0.0000000000000000 0.0000000000000000 0.0000000000000000 | 0.0000000000000000 0.0000000000000000 0.0000000000000000 | ||
0.2500000000000000 0.2500000000000000 0.2500000000000000 | 0.2500000000000000 0.2500000000000000 0.2500000000000000 | ||
This is the complete {{FILE|POSCAR}} file: | |||
Zincblende GaAs | Zincblende GaAs | ||
5.65000000000 | 5.65000000000 | ||
| Line 67: | Line 85: | ||
If you have access to {{py4vasp}}, the structure can be visualized with two lines of Python code in a Jupyter notebook. | If you have access to {{py4vasp}}, the structure can be visualized with two lines of Python code in a Jupyter notebook. | ||
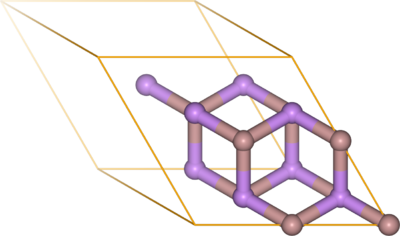
[[File:GaAs_zincblende_py4vasp_2SC.png|400px|thumb|riight|Visualization of the POSCAR with py4vasp]] | [[File:GaAs_zincblende_py4vasp_2SC.png|400px|thumb|riight|Visualization of the {{FILE|POSCAR}} file of GaAs with {{py4vasp}}.]] | ||
<syntaxhighlight lang="python"> | <syntaxhighlight lang="python"> | ||
from py4vasp import calculation | from py4vasp import calculation | ||
| Line 80: | Line 98: | ||
from ase.io.vasp import write_vasp | from ase.io.vasp import write_vasp | ||
atoms = bulk("GaAs", crystalstructure="zincblende", a=5.65) | atoms = bulk("GaAs", crystalstructure="zincblende", a=5.65) | ||
write_vasp("POSCAR", atoms, direct=True, sort=False) | write_vasp("POSCAR", atoms, direct=True, sort=False) | ||
</syntaxhighlight> | </syntaxhighlight> | ||
=== Creating the POTCAR file === | === Creating the {{FILE|POTCAR}} file === | ||
We have already decided to use {{TAGO|XC|CA}}, and can create the {{FILE|POTCAR}} file as discussed on the [[Preparing_a_POTCAR| preparing a POTCAR]] page. | We have already decided to use {{TAGO|XC|CA}}, and can create the {{FILE|POTCAR}} file as discussed on the [[Preparing_a_POTCAR| preparing a POTCAR]] page. | ||
=== Creating the KPOINTS file === | === Creating the {{FILE|KPOINTS}} file === | ||
Since our structure is face-centered cubic, we create a [[KPOINTS#Regular_k-point_mesh| regular Gamma-centered]] '''k'''-point mesh according to the [[KPOINTS#Symmetry_reduction_of_the_mesh| symmetry considerations]] for | Since our structure is face-centered cubic, we create a [[KPOINTS#Regular_k-point_mesh| regular Gamma-centered]] '''k'''-point mesh according to the [[KPOINTS#Symmetry_reduction_of_the_mesh| symmetry considerations]] for | ||
{{FILE| KPOINTS}} files | {{FILE| KPOINTS}} files. | ||
Regular k-point mesh | Regular k-point mesh | ||
0 | 0 | ||
| Line 99: | Line 115: | ||
7 7 7 | 7 7 7 | ||
=== Creating the INCAR file === | === Creating the {{FILE|INCAR}} file === | ||
We | We chose the efficient combination of a [[blocked-Davidson algorithm]] and the [[RMM-DIIS]] algorithm which can be selected with {{TAGO|ALGO|Fast}}. | ||
ALGO = Fast | {{TAGBL|ALGO}} = Fast | ||
GaAs is a semiconductor so we could use the tetrahedron method {{TAGO|ISMEAR|-5}} | GaAs is a semiconductor, so we could use the tetrahedron method {{TAGO|ISMEAR|-5}}, but bandgaps are underestimated systematically by DFT and XC functional or lattice parameter may fail to reproduce experimental results. Thus, following the recommendation on [[smearing technique#Which method to use |electronic smearing techniques]], it is safer to select Gaussian smearing and a small smearing width: | ||
ISMEAR = 0 | {{TAGBL|ISMEAR}} = 0 | ||
SIGMA = 0. | {{TAGBL|SIGMA}} = 0.1 | ||
For an initial guess of the plane-wave cutoff energy {{TAG| ENCUT}} we can search for ENMAX in | {{TAGBL|EFERMI}} = MIDGAP | ||
ENCUT = 285 | For an initial guess of the plane-wave cutoff energy {{TAG| ENCUT}}, we can search for {{TAG|ENMAX}} in the {{FILE|POTCAR}}, e.g. by <code>grep ENMAX POTCAR</code>, and set the largest as a starting point. In preceding calculations this value should be increased, e.g. by increments of approximately 20%. Accordingly, in the first run we set: | ||
For the break condition of the self-consistent loop we select <math>1\times10^{-6}</math> eV: | {{TAGBL|ENCUT}} = 285 | ||
EDIFF = 1.0E-06 | For the break condition of the self-consistent loop, we select <math>1\times10^{-6}</math> eV: | ||
The complete {{FILE| INCAR}} file | {{TAGBL|EDIFF}} = 1.0E-06 | ||
ALGO = Fast | The complete {{FILE| INCAR}} file is: | ||
ISMEAR = 0 | {{TAGBL|XC}} = CA | ||
SIGMA = 0. | {{TAGBL|ALGO}} = Fast | ||
ENCUT = 285 | {{TAGBL|ISMEAR}} = 0 | ||
EDIFF = 1.0E-06 | {{TAGBL|EFERMI}} = MIDGAP | ||
{{TAGBL|SIGMA}} = 0.1 | |||
{{TAGBL|ENCUT}} = 285 | |||
{{TAGBL|EDIFF}} = 1.0E-06 | |||
=== | === Performing a dryrun === | ||
We are not doing a [[:Category:Noncollinear_magnetism| noncollinear]], nor a Gamma-only calculation, thus we execute a VASP [[Command-line_arguments#--dry-run_/_-n | dry-run]] with the standard executable: | We are not doing a [[:Category:Noncollinear_magnetism| noncollinear]], nor a Gamma-only calculation, thus we execute a VASP [[Command-line_arguments#--dry-run_/_-n | dry-run]] with the standard executable: | ||
/your/vasp_dir/bin/vasp_std --dry-run | /your/vasp_dir/bin/vasp_std --dry-run | ||
Which will print a warning about the [[Command-line_arguments#--dry-run_/_-n | dry-run]] and some information about the MPI-ranks, | Which will print a warning about the [[Command-line_arguments#--dry-run_/_-n | dry-run]] and some information about the MPI-ranks, OMP-threads, the VASP version, and the input structure. Mistakes in the setup, e.g. if the order of elements in the {{FILE| POSCAR}} and {{FILE| POTCAR}} do not match, warnings are printed. | ||
We can now check the {{FILE| OUTCAR}} file and find the total number of '''k'''-points, 20, and | We can now check the {{FILE| OUTCAR}} file and find the total number of '''k'''-points, 20, and number of bands ({{TAG|NBANDS}}), 13. This means a relatively low number of bands and a decent number of '''k''' points. If we want to run our calculation on 4 MPI ranks, setting {{TAGO|KPAR|4}} is an excellent choice for parallelization. Mind that the [[NBANDS#Parallelization|parallelization changes number of bands]]. | ||
=== Running the calculation === | === Running the calculation === | ||
| Line 132: | Line 151: | ||
distrk: each k-point on 1 cores, 4 groups | distrk: each k-point on 1 cores, 4 groups | ||
distr: one band on 1 cores, 1 groups | distr: one band on 1 cores, 1 groups | ||
vasp.6.5. | vasp.6.5.0 16Dec24 (build Dec 18 2024 11:18:52) complex | ||
POSCAR found type information on POSCAR GaAs | POSCAR found type information on POSCAR GaAs | ||
POSCAR found : 2 types and 2 ions | POSCAR found : 2 types and 2 ions | ||
Reading from existing POTCAR | |||
scaLAPACK will be used | scaLAPACK will be used | ||
Reading from existing POTCAR | |||
LDA part: xc-table for (Slater(with rela. corr.)+CA(PZ)) | LDA part: xc-table for (Slater(with rela. corr.)+CA(PZ)) | ||
, standard interpolation | , standard interpolation | ||
| Line 146: | Line 167: | ||
entering main loop | entering main loop | ||
N E dE d eps ncg rms rms(c) | N E dE d eps ncg rms rms(c) | ||
DAV: 1 0. | DAV: 1 0.623500523606E+02 0.62350E+02 -0.70852E+03 528 0.135E+03 | ||
DAV: 2 -0. | DAV: 2 -0.533903918847E+01 -0.67689E+02 -0.65331E+02 580 0.246E+02 | ||
DAV: 3 -0. | DAV: 3 -0.978648308483E+01 -0.44474E+01 -0.44252E+01 635 0.613E+01 | ||
DAV: 4 -0. | DAV: 4 -0.985351010991E+01 -0.67027E-01 -0.67012E-01 614 0.819E+00 | ||
DAV: 5 -0. | DAV: 5 -0.985490478939E+01 -0.13947E-02 -0.13947E-02 641 0.931E-01 0.301E+00 | ||
RMM: 6 -0. | RMM: 6 -0.966994813504E+01 0.18496E+00 -0.21049E-01 715 0.453E+00 0.175E+00 | ||
RMM: 7 -0. | RMM: 7 -0.962995486843E+01 0.39993E-01 -0.10315E-01 701 0.182E+00 0.574E-01 | ||
RMM: 8 -0. | RMM: 8 -0.962647867206E+01 0.34762E-02 -0.12692E-02 740 0.127E+00 0.937E-02 | ||
RMM: 9 -0. | RMM: 9 -0.962642442346E+01 0.54249E-04 -0.21087E-03 757 0.536E-01 0.594E-02 | ||
RMM: 10 -0. | RMM: 10 -0.962647797834E+01 -0.53555E-04 -0.39237E-04 794 0.212E-01 0.167E-02 | ||
RMM: 11 -0. | RMM: 11 -0.962646653288E+01 0.11445E-04 -0.91747E-05 785 0.105E-01 0.529E-03 | ||
RMM: 12 -0. | RMM: 12 -0.962646808711E+01 -0.15542E-05 -0.17691E-05 735 0.426E-02 0.300E-03 | ||
RMM: 13 -0. | RMM: 13 -0.962646810096E+01 -0.13852E-07 -0.27033E-06 491 0.223E-02 | ||
1 F= -. | 1 F= -.96264681E+01 E0= -.96264536E+01 d E =-.289650E-04 | ||
writing wavefunctions | writing wavefunctions | ||
You can now use the [[:Category:Output_files| output files]] to analyze the [[:Category:Electronic_ground-state_properties| electronic ground-state properties]]. Do not forget to perform a convergence study before reporting a value, see [[#Step 9|Step 9]]. | |||
==Related tags and articles== | ==Related tags and articles== | ||
{{FILE| INCAR}}, {{FILE| POSCAR}}, {{FILE| KPOINTS}}, {{FILE| POTCAR}}, | [[:Category:Input files | Input files]]: {{FILE| INCAR}}, {{FILE| POSCAR}}, {{FILE| KPOINTS}}, {{FILE| POTCAR}}, | ||
[[:Category:Parallelization| Parallelization]] | |||
[[:Category:Output_files| Output files]] and [[:Category:Electronic ground-state properties| Electronic ground-state properties]] | |||
== References == | == References == | ||
[[Category:Howto|Howto]] [[Category:Calculation_setup|Calculation setup]] [[Category:Electronic minimization|Electronic minmization]] | |||
Latest revision as of 13:29, 11 March 2025
Setting up an electronic minimization calculation using density-functional theory requires a few steps. The input files must be created or copied into the execution folder. This includes making a few choices for the k-point sampling and electronic smearing, minimization algorithm, and exchange-correlation functionals. A dry-run can be used to review settings and select appropriate parallelization tags. After running the calculation, the output can be analyzed.
Step-by-step instructions
Create the input files
Step 1: Create a POSCAR file containing the structure for which you want to compute the electronic groundstate. External tools like VESTA[1], or Python packages like the Atomic Simulation Environment (ASE)[2] or pymatgen[3] can help with this step.
Step 2: Choose an exchange-correlation (XC) functional appropriate for your material and quantity of interest.
Step 3: Create a suitable POTCAR file by following the instructions on our preparing a POTCAR page.
Step 4: Create a KPOINTS file to define the integration mesh in reciprocal space. Including a single k point at the origin, i.e. the Gamma point, neglects all interactions beyond the unit cell. This is appropriate for isolates systems like a single molecule or in large supercells. For bulk systems, start with a regular mesh. For shorter lattice vectors, more k points are required to achieve the same sampling density. Consult the symmetry reduction section of the KPOINTS page to select the appropriate mesh type. Alternatively to a KPOINTS file, the KSPACING can be used.
Step 5: Write an INCAR file. It is recommended to start from a rather minimal file, and only specify the most important tags:
- XC to specify the exchange-correlation functional.
- ALGO to select the algorithm for electronic minimization.
- ISMEAR to select the type of electronic smearing technique.
- SIGMA to choose an appropriate smearing width of the electronic smearing.
- ENCUT to set the plane-wave energy cutoff.
- EDIFF to specify the global break condition for the electronic self-consistent loop
Optimize your settings
Step 6 (optional): Select the appropriate version of the VASP executable. I.e. vasp_gam if you only want to use the Gamma point for reciprocal space integration, vasp_ncl for noncollinear magnetic calculations, or vasp_std for anything else. Then Run a dry-run calculation to validate settings and uncover possible errors. Open a terminal, go to the calculation directory that contains all input files and run
/path/to/your/vasp_std --dry-run
Step 7 (optional): Inspect the OUTCAR file of your dry-run. Take note of the number of bands, NBANDS, and the number of k-points, NKPTS, especially. Follow the guidelines on the optimizing the parallelization page to set NCORE and/or KPAR in the INCAR file.
Run the calculation
Step 8: Run the calculation. If you are new to VASP, or unsure about the calculation setup, run a small calculation and monitor the screen output. For parallel execution on 4 MPI ranks, the command reads
mpirun -np 4 /path/to/your/vasp_std
At an HPC center, submit your job with a submission script. Ask your system administrator for help.
Once the calculation is finished, you have access to the electronic ground-state properties via the output files for the selected parameters. Check the OUTCAR file for warnings or advice. For help, consult the page about troubleshooting electronic convergence and search our Forum for similar issues.
Recommendations and advice
| Mind: Make sure to specify the lattice vectors and ionic positions in the POSCAR with at least 7 digits of precision to ensure the symmetry analysis can function accurately. |
Tips
|
Example
We will do a small DFT calculation of GaAs in the zincblende structure, using the local-density approximation (LDA) with the Perdew-Zunger parametrization of Ceperley-Alder Monte Carlo correlation data.[4][5]. Thus, our XC functional will be set to XC = CA.
Setting up the POSCAR file
The POSCAR file starts with a comment line and a scaling factor, which in our case corresponds to the lattice parameter of GaAs, around 5.65 Angstrom.
Zincblende GaAs 5.65000000000
Next we need to define the lattice vectors. Zincblende is a face-centered cubic (fcc) structure with two different elements in the unit cell. We can describe the fcc lattice with three vectors, pointing from the origin to the face-centers of the cube:
0.0000000000000000 0.5000000000000000 0.5000000000000000
0.5000000000000000 0.0000000000000000 0.5000000000000000
0.5000000000000000 0.5000000000000000 0.0000000000000000
Now, we define the ion types, and in the line below the number of ions in the structure for each type:
Ga As 1 1
Specify the positions of the atoms in direct coordinates, with Ga at the origin and As a quarter along the diagonal of the cube:
Direct 0.0000000000000000 0.0000000000000000 0.0000000000000000 0.2500000000000000 0.2500000000000000 0.2500000000000000
This is the complete POSCAR file:
Zincblende GaAs
5.65000000000
0.0000000000000000 0.5000000000000000 0.5000000000000000
0.5000000000000000 0.0000000000000000 0.5000000000000000
0.5000000000000000 0.5000000000000000 0.0000000000000000
Ga As
1 1
Direct
0.0000000000000000 0.0000000000000000 0.0000000000000000
0.2500000000000000 0.2500000000000000 0.2500000000000000
If you have access to py4vasp, the structure can be visualized with two lines of Python code in a Jupyter notebook.
from py4vasp import calculation
calculation.structure.plot(supercell=2,selection="POSCAR")
If ASE[2] is installed, an equivalent POSCAR file can be created as follows:
from ase.build import bulk
from ase.io.vasp import write_vasp
atoms = bulk("GaAs", crystalstructure="zincblende", a=5.65)
write_vasp("POSCAR", atoms, direct=True, sort=False)
Creating the POTCAR file
We have already decided to use XC = CA, and can create the POTCAR file as discussed on the preparing a POTCAR page.
Creating the KPOINTS file
Since our structure is face-centered cubic, we create a regular Gamma-centered k-point mesh according to the symmetry considerations for KPOINTS files.
Regular k-point mesh 0 Gamma 7 7 7
Creating the INCAR file
We chose the efficient combination of a blocked-Davidson algorithm and the RMM-DIIS algorithm which can be selected with ALGO = Fast.
ALGO = Fast
GaAs is a semiconductor, so we could use the tetrahedron method ISMEAR = -5, but bandgaps are underestimated systematically by DFT and XC functional or lattice parameter may fail to reproduce experimental results. Thus, following the recommendation on electronic smearing techniques, it is safer to select Gaussian smearing and a small smearing width:
ISMEAR = 0 SIGMA = 0.1 EFERMI = MIDGAP
For an initial guess of the plane-wave cutoff energy ENCUT, we can search for ENMAX in the POTCAR, e.g. by grep ENMAX POTCAR, and set the largest as a starting point. In preceding calculations this value should be increased, e.g. by increments of approximately 20%. Accordingly, in the first run we set:
ENCUT = 285
For the break condition of the self-consistent loop, we select eV:
EDIFF = 1.0E-06
The complete INCAR file is:
XC = CA ALGO = Fast ISMEAR = 0 EFERMI = MIDGAP SIGMA = 0.1 ENCUT = 285 EDIFF = 1.0E-06
Performing a dryrun
We are not doing a noncollinear, nor a Gamma-only calculation, thus we execute a VASP dry-run with the standard executable:
/your/vasp_dir/bin/vasp_std --dry-run
Which will print a warning about the dry-run and some information about the MPI-ranks, OMP-threads, the VASP version, and the input structure. Mistakes in the setup, e.g. if the order of elements in the POSCAR and POTCAR do not match, warnings are printed.
We can now check the OUTCAR file and find the total number of k-points, 20, and number of bands (NBANDS), 13. This means a relatively low number of bands and a decent number of k points. If we want to run our calculation on 4 MPI ranks, setting KPAR = 4 is an excellent choice for parallelization. Mind that the parallelization changes number of bands.
Running the calculation
After adding KPAR = 4 to the INCAR file, we run the calculation on 4 MPI ranks:
mpirun -np 4 /your/vasp_dir/bin/vasp_std
Consult the page on screen output for details about the information VASP prints out. For this example it should be similar to this:
running 4 mpi-ranks, with 1 threads/rank, on 1 nodes
distrk: each k-point on 1 cores, 4 groups
distr: one band on 1 cores, 1 groups
vasp.6.5.0 16Dec24 (build Dec 18 2024 11:18:52) complex
POSCAR found type information on POSCAR GaAs
POSCAR found : 2 types and 2 ions
Reading from existing POTCAR
scaLAPACK will be used
Reading from existing POTCAR
LDA part: xc-table for (Slater(with rela. corr.)+CA(PZ))
, standard interpolation
POSCAR, INCAR and KPOINTS ok, starting setup
FFT: planning ... GRIDC
FFT: planning ... GRID_SOFT
FFT: planning ... GRID
WAVECAR not read
entering main loop
N E dE d eps ncg rms rms(c)
DAV: 1 0.623500523606E+02 0.62350E+02 -0.70852E+03 528 0.135E+03
DAV: 2 -0.533903918847E+01 -0.67689E+02 -0.65331E+02 580 0.246E+02
DAV: 3 -0.978648308483E+01 -0.44474E+01 -0.44252E+01 635 0.613E+01
DAV: 4 -0.985351010991E+01 -0.67027E-01 -0.67012E-01 614 0.819E+00
DAV: 5 -0.985490478939E+01 -0.13947E-02 -0.13947E-02 641 0.931E-01 0.301E+00
RMM: 6 -0.966994813504E+01 0.18496E+00 -0.21049E-01 715 0.453E+00 0.175E+00
RMM: 7 -0.962995486843E+01 0.39993E-01 -0.10315E-01 701 0.182E+00 0.574E-01
RMM: 8 -0.962647867206E+01 0.34762E-02 -0.12692E-02 740 0.127E+00 0.937E-02
RMM: 9 -0.962642442346E+01 0.54249E-04 -0.21087E-03 757 0.536E-01 0.594E-02
RMM: 10 -0.962647797834E+01 -0.53555E-04 -0.39237E-04 794 0.212E-01 0.167E-02
RMM: 11 -0.962646653288E+01 0.11445E-04 -0.91747E-05 785 0.105E-01 0.529E-03
RMM: 12 -0.962646808711E+01 -0.15542E-05 -0.17691E-05 735 0.426E-02 0.300E-03
RMM: 13 -0.962646810096E+01 -0.13852E-07 -0.27033E-06 491 0.223E-02
1 F= -.96264681E+01 E0= -.96264536E+01 d E =-.289650E-04
writing wavefunctions
You can now use the output files to analyze the electronic ground-state properties. Do not forget to perform a convergence study before reporting a value, see Step 9.
Related tags and articles
Input files: INCAR, POSCAR, KPOINTS, POTCAR,
Output files and Electronic ground-state properties
